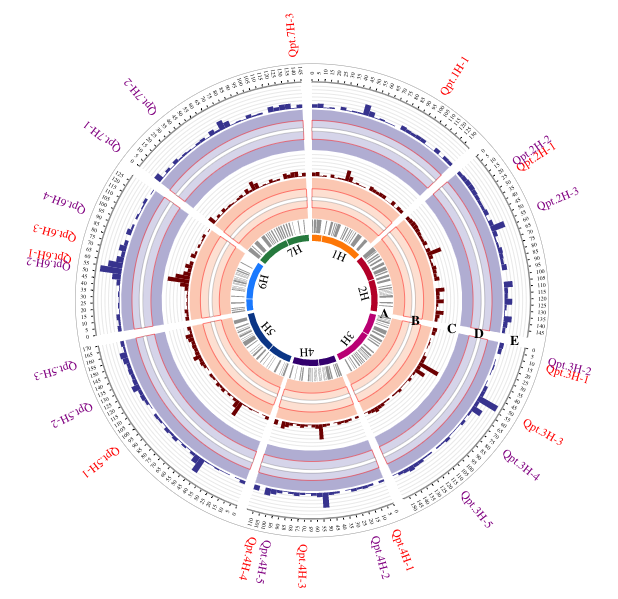
An interactive Circos plot indicating QTL involved in net blotch resistance and displayed using histogram, arc, link, background, text function.
RCircos is a simple and flexible R package for generating 2D Circos plot, which includes heatmap, histogram, lines, scatter and other types of plot.
In order to illustrate the simplicity of interacCircos and help users porting their existing RCircos code to interacCircos, we compared the generation of a multi-track Circos plot using RCircos and interacCircos.
The full scripts for both package are available here: RCircos-demo.R or interacCircos-demo.R
The step by step comparison is displayed below:
library(RCircos)
# Load hg19 and draw ideogram in RCircos
data(UCSC.HG19.Human.CytoBandIdeogram)
RCircos.Set.Core.Components(cyto.info=UCSC.HG19.Human.CytoBandIdeogram,
chr.exclude=NULL,tracks.inside=5,tracks.outside=0)
RCircos.Set.Plot.Area()
RCircos.Chromosome.Ideogram.Plot()
library(interacCircos)
# The hg19 reference is already embedded in interacCircos,
# You can directly load hg19 by using 'genome' parameters,
# and load ideogram for hg19 by hg19_ideogram
# Customize ideogram track and its animation
ideogram_track<-CircosArc("arc01",data=hg19_ideogram,
innerRadius = 210,outerRadius = 230,animationDisplay = TRUE,
animationDelay=10,animationTime = 100)
# Generate chromosome track and ideogram track
Circos(genome="hg19",moduleList=ideogram_track,innerRadius = 238,
outerRadius = 240)
Note that we extracted the scatter data of RCircos for interacCircos. The data processing script for scatter data is available here, the extracted data is available here.
library(RCircos)
# Load hg19 and draw ideogram in RCircos
data(UCSC.HG19.Human.CytoBandIdeogram)
RCircos.Set.Core.Components(cyto.info=UCSC.HG19.Human.CytoBandIdeogram,
chr.exclude=NULL,tracks.inside=5,tracks.outside=0)
RCircos.Set.Plot.Area()
RCircos.Chromosome.Ideogram.Plot()
#Load scatter data
data(RCircos.Scatter.Data)
# Add scatter track into plot
RCircos.Scatter.Plot(RCircos.Scatter.Data, data.col=5,
track.num=1, side='in', by.fold=1)
llibrary(interacCircos)
# The hg19 reference is already embedded in interacCircos,
# You can directly load hg19 by using 'genome' parameters,
# and load ideogram for hg19 by hg19_ideogram
# Customize ideogram track and its animation
ideogram_track<-CircosArc("arc01",data=hg19_ideogram,
innerRadius = 210,outerRadius = 230,
animationDisplay = TRUE,
animationDelay=10,animationTime = 100)
# Load scatter data
load("scatterData.rda")
# The snp track and its background
# The SNP function of interacCircos is same as scatter of RCircos
snp_track<-CircosSnp("snp01",data=scatterData,minRadius = 183,
maxRadius = 205,animationDisplay = TRUE,
animationDelay = 0,animationTime = 2000,
animationType = "linear",circleSize = 1)
bg_track1<-CircosBackground("bg01",axisShow = TRUE,minRadius = 183,
maxRadius = 205,fillColors ="rgb(241,220,179)",
animationDisplay = TRUE,animationType = "linear")
# Generate chromosome track and ideogram track
Circos(genome="hg19",moduleList=ideogram_track+snp_track+bg_track1,
innerRadius = 238,outerRadius = 240,SNPMouseOverDisplay = TRUE,
SNPMouseOverTooltipsSetting = "style1",SNPMouseOutDisplay = TRUE,SNPMouseOutColor = NULL)
Note that we extracted the histogram data of RCircos for interacCircos. The data processing script for histogram data is available here, the extracted data is available here.
library(RCircos)
# Load hg19 and draw ideogram in RCircos
data(UCSC.HG19.Human.CytoBandIdeogram)
RCircos.Set.Core.Components(cyto.info=UCSC.HG19.Human.CytoBandIdeogram,
chr.exclude=NULL,tracks.inside=5,tracks.outside=0)
RCircos.Set.Plot.Area()
RCircos.Chromosome.Ideogram.Plot()
# Load data for scatter track
data(RCircos.Scatter.Data)
# Add scatter track into plot
RCircos.Scatter.Plot(RCircos.Scatter.Data, data.col=5,track.num=1,
side='in', by.fold=1)
# Load data for histogram track
data(RCircos.Histogram.Data)
# Add histogram track into plot
RCircos.Histogram.Plot(RCircos.Histogram.Data,data.col = 4,
track.num = 3,side = 'in')
library(interacCircos)
# The hg19 reference is already embedded in interacCircos,
# You can directly load hg19 by using 'genome' parameters,
# and load ideogram for hg19 by hg19_ideogram
# Customize ideogram track and its animation
ideogram_track<-CircosArc("arc01",data=hg19_ideogram,
innerRadius = 210,outerRadius = 230,
animationDisplay = TRUE,
animationDelay=10,animationTime = 100)
# Load scatter data
load("scatterData.rda")
# The snp track and its background
# The SNP function of interacCircos is same as scatter of RCircos
snp_track<-CircosSnp("snp01",data=scatterData,minRadius = 183,
maxRadius = 205,animationDisplay = TRUE,animationDelay = 0,
animationTime = 2000,animationType = "linear",circleSize = 1)
bg_track1<-CircosBackground("bg01",axisShow = TRUE,minRadius = 183,maxRadius = 205,
fillColors ="rgb(241,220,179)",animationDisplay = TRUE,animationType = "linear")
# Load histogram data
load("histogramData.rda")
# The histogram track
histogram_track<-CircosHistogram("histogram01",data=histogramData,
minRadius = 143,maxRadius = 165,animationDisplay = TRUE)
bg_track2<-CircosBackground("bg02",axisShow = TRUE,minRadius = 143,
maxRadius = 165,fillColors ="rgb(241,220,179)",
animationDisplay = TRUE,animationType = "linear")
# Generate chromosome track and ideogram track
Circos(genome="hg19",moduleList=ideogram_track+snp_track+bg_track1+
histogram_track+bg_track2,innerRadius = 238,outerRadius = 240,
SNPMouseOverDisplay = TRUE,SNPMouseOverTooltipsSetting = "style1",
SNPMouseOutDisplay = TRUE,SNPMouseOutColor = NULL,
HISTOGRAMMouseOverDisplay = TRUE,HISTOGRAMMouseOverTooltipsSetting = "style1",
HISTOGRAMMouseOutDisplay = TRUE,HISTOGRAMMouseOutColor = NULL)
Note that we extracted the link data of RCircos for interacCircos. The data processing script for link data is available here, the extracted data is available here.
library(RCircos)
# Load hg19 and draw ideogram in RCircos
data(UCSC.HG19.Human.CytoBandIdeogram)
RCircos.Set.Core.Components(cyto.info=UCSC.HG19.Human.CytoBandIdeogram,
chr.exclude=NULL,tracks.inside=5,tracks.outside=0)
RCircos.Set.Plot.Area()
RCircos.Chromosome.Ideogram.Plot()
# Load data for scatter track
data(RCircos.Scatter.Data)
# Add scatter track into plot
RCircos.Scatter.Plot(RCircos.Scatter.Data, data.col=5,track.num=1,
side='in', by.fold=1)
# Load data for histogram track
data(RCircos.Histogram.Data)
# Add histogram track into plot
RCircos.Histogram.Plot(RCircos.Histogram.Data,data.col = 4,track.num = 3,
side = 'in')
# Load data for link track
data(RCircos.Link.Data)
# Add link track into plot
RCircos.Link.Plot(RCircos.Link.Data,track.num=4,TRUE)
library(interacCircos)
# The hg19 reference is already embedded in interacCircos,
# You can directly load hg19 by using 'genome' parameters,
# and load ideogram for hg19 by hg19_ideogram
# Customize ideogram track and its animation
ideogram_track<-CircosArc("arc01",data=hg19_ideogram,
innerRadius = 210,outerRadius = 230,animationDisplay = TRUE,
animationDelay=10,animationTime = 100)
# Load scatter data
load("scatterData.rda")
# The snp track and its background
# The SNP function of interacCircos is same as scatter of RCircos
snp_track<-CircosSnp("snp01",data=scatterData,minRadius = 183,maxRadius = 205,
animationDisplay = TRUE,animationDelay = 0,animationTime = 2000,
animationType = "linear",circleSize = 1)
bg_track1<-CircosBackground("bg01",axisShow = TRUE,minRadius = 183,maxRadius = 205,
fillColors ="rgb(241,220,179)",animationDisplay = TRUE,animationType = "linear")
# Load histogram data
load("histogramData.rda")
# The histogram track
histogram_track<-CircosHistogram("histogram01",data=histogramData,minRadius = 143,
maxRadius = 165,animationDisplay = TRUE)
bg_track2<-CircosBackground("bg02",axisShow = TRUE,minRadius = 143,maxRadius = 165,
fillColors ="rgb(241,220,179)",
animationDisplay = TRUE,animationType = "linear")
# Load link data
load("linkData.rda")
# The link track
link_track01<-CircosLink("link01",data=linkData[which(linkData$g1chr==linkData$g2chr),],
radius = 140,width = 1,fillColor = "red",animationDisplay =TRUE,
animationTime = 4000,animationDelay = 200)
link_track02<-CircosLink("link02",data=linkData[which(!linkData$g1chr==linkData$g2chr),],
radius = 140,width = 1,fillColor = "blue",animationDisplay = TRUE,
animationTime = 4000,animationDelay = 200)
# Generate chromosome track and ideogram track
Circos(genome="hg19",moduleList=ideogram_track+snp_track+bg_track1+histogram_track+
bg_track2+link_track01+link_track02,innerRadius = 238,outerRadius = 240,
SNPMouseOverDisplay = TRUE,SNPMouseOverTooltipsSetting = "style1",
SNPMouseOutDisplay = TRUE,SNPMouseOutColor = NULL,
HISTOGRAMMouseOverDisplay = TRUE,HISTOGRAMMouseOverTooltipsSetting = "style1",
HISTOGRAMMouseOutDisplay = TRUE,HISTOGRAMMouseOutColor = NULL)