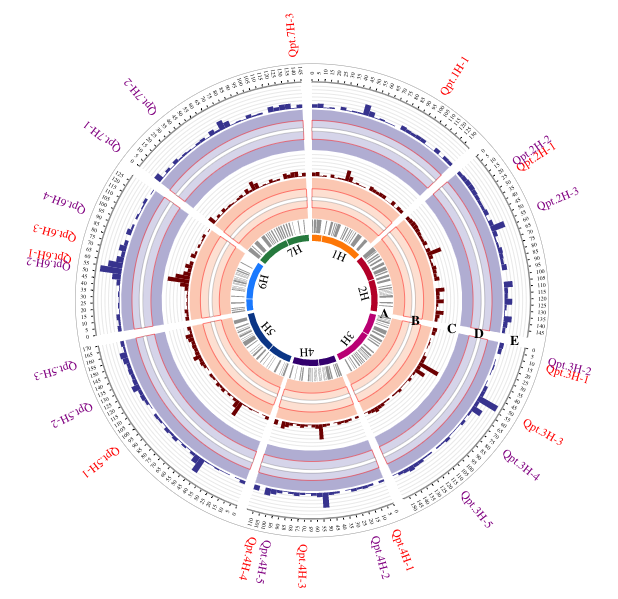
An interactive Circos plot indicating QTL involved in net blotch resistance and displayed using histogram, arc, link, background, text function.
Normally, the SNP data is stored in .vcf format, thus we use an real VCF file as example here.
The example of real SNP file is available here
First, we need to extract the effective SNP columns from the original VCF file
# Load original SNP data
library(interacCircos)
library(stringr)
SNP_data<-read.table("snpExample_random_from_1000G.vcf",sep = "\t")
SNP_data<-SNP_data[,c(1,2,8)]
colnames(SNP_data)<-c("chr","pos","value")
SNP_data$value<-as.integer(str_split_fixed(str_split_fixed(SNP_data$value,
";", 2)[,1],"=",2)[,2])
After processing, the SNP_data should looks like below:
| chr | pos | value |
|---|---|---|
| 1 | 70573281 | 9 |
| 1 | 167102542 | 1 |
| 1 | 238165263 | 1 |
| 2 | 52714177 | 1827 |
| ... | ... | ... |
In order to load other functional tracks, users should first construct a chromosome track!
# Load original SNP data
library(interacCircos)
library(stringr)
SNP_data<-read.table("snpExample_random_from_1000G.vcf",sep = "\t")
SNP_data<-SNP_data[,c(1,2,8)]
colnames(SNP_data)<-c("chr","pos","value")
SNP_data$value<-as.integer(str_split_fixed(str_split_fixed(SNP_data$value,
";", 2)[,1],"=",2)[,2])
Circos()
In order to add snp track, users should first constrct a CircosSNP() function and then load this track on Circos() function
# Load original SNP data
library(interacCircos)
library(stringr)
SNP_data<-read.table("snpExample_random_from_1000G.vcf",sep = "\t")
SNP_data<-SNP_data[,c(1,2,8)]
colnames(SNP_data)<-c("chr","pos","value")
SNP_data$value<-as.integer(str_split_fixed(str_split_fixed(SNP_data$value, ";", 2)[,1],"=",2)[,2])
SNP_track<-CircosSnp('SNP01', minRadius =150, maxRadius = 190, data = SNP_data,fillColor= "#9ACD32",
circleSize= 2)
Circos(moduleList = SNP_track)
It's not pretty enough when only add a snp track, users can add other tracks to increase readability.
# Load original SNP data
library(interacCircos)
library(stringr)
SNP_data<-read.table("snpExample_random_from_1000G.vcf",sep = "\t")
SNP_data<-SNP_data[,c(1,2,8)]
colnames(SNP_data)<-c("chr","pos","value")
SNP_data$value<-as.integer(str_split_fixed(str_split_fixed(SNP_data$value, ";", 2)[,1],"=",2)[,2])
SNP_track<-CircosSnp('SNP01', minRadius =150, maxRadius = 190, data = SNP_data,fillColor= "#9ACD32",
circleSize= 2)
Bg_track<-CircosBackground('BG01',minRadius = 145, maxRadius = 200)
Circos(moduleList = SNP_track+Bg_track)
As you know, interacCircos is interactive as it can add animation and mouse event to plot. Here, we add a mouse move event and a opening animation for snp track
# Load original SNP data
library(interacCircos)
library(stringr)
SNP_data<-read.table("snpExample_random_from_1000G.vcf",sep = "\t")
SNP_data<-SNP_data[,c(1,2,8)]
colnames(SNP_data)<-c("chr","pos","value")
SNP_data$value<-as.integer(str_split_fixed(str_split_fixed(SNP_data$value, ";", 2)[,1],"=",2)[,2])
SNP_track<-CircosSnp('SNP01', minRadius =150, maxRadius = 190, data = SNP_data,fillColor= "#9ACD32",
circleSize= 2, animationDisplay =TRUE,animationTime= 2000,animationDelay= 0,
animationType= "linear")
Bg_track<-CircosBackground('BG01',minRadius = 145, maxRadius = 200)
Circos(moduleList = SNP_track+Bg_track,SNPMouseEvent = TRUE,SNPMouseOverDisplay = TRUE,
SNPMouseOverTooltipsSetting = "style1",SNPMouseOutDisplay = TRUE,SNPMouseOutColor = NULL)