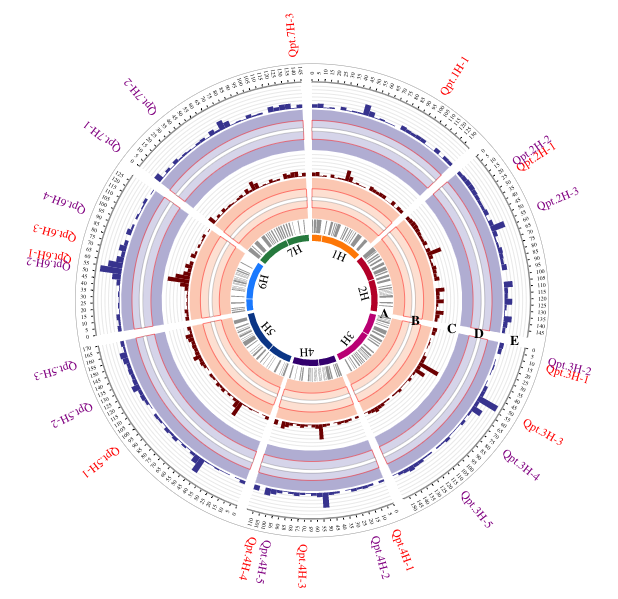
An interactive Circos plot indicating QTL involved in net blotch resistance and displayed using histogram, arc, link, background, text function.
CircosArc() can display the CNV without value, Gene domain, Chromosome band, etc.
library(interacCircos)
arcData=arcExample
Circos(moduleList=CircosArc('Arc01', outerRadius = 212,
innerRadius = 224, data=arcData,animationDisplay = TRUE),
genome=list("EGFR"=1211),
outerRadius = 220,genomeFillColor = c("grey"),
ARCMouseOverDisplay = TRUE,ARCMouseOverTooltipsSetting = "style1",
ARCMouseOutDisplay = TRUE,ARCMouseOutColor = NULL)
Note : No data input for CircosArc(). The details for parameters are available by ?CircosArc().
CircosAuxLine() can display Auxiliary line for better explain the relationship between modules.
library(interacCircos)
Circos(moduleList=CircosAuxLine('AuxLine01',
animationDisplay = TRUE,animationType = "linear",
animationDelay =0,animationTime = 1000))
Note : No data input for CircosAuxLine(). The details for parameters are available by ?CircosAuxLine().
CircosBackground() can display background and axis circles for other functions.
library(interacCircos)
Circos(moduleList=CircosBackground('bg01', fillColors="#FFEEEE",
borderSize = 1,animationDisplay = TRUE))
Note : No data input for CircosBackground(). The details for parameters are available by ?CircosBackground().
CircosBubble() can display the gene expression data as well as its classification, etc.
library(interacCircos)
bubbleData=bubbleExample
Circos(moduleList=CircosBubble('Bubble01', maxRadius = 230,
minRadius = 170, data=bubbleData, blockStroke = TRUE, bubbleMaxSize =10,
bubbleMinSize = 2, maxColor = "red", minColor = "yellow", totalLayer =3,
animationDisplay = TRUE, animationType="linear"),
genome = list("2L"=23011544,"2R"=21146708,
"3L"=24543557,"3R"= 27905053,"X"=22422827,"4"=1351857),
BUBBLEMouseOverDisplay =TRUE,BUBBLEMouseOverTooltipsSetting = "style1",
innerRadius = 236,BUBBLEMouseOutDisplay = TRUE,BUBBLEMouseOutColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosBubble().
CircosChord() can display the relationship between gene, exon, protein, etc. (Using matrix as chord path)
library(interacCircos)
chordData=chordExample
Circos(moduleList=CircosChord('CHORD01', data = chordData,innerRadius= 210,outerRadius= 211,fillOpacity=0.67,
strokeColor="black",strokeWidth= "1px",outerARCText=FALSE,autoFillColor = FALSE,
fillColor = c("blue","green","yellow","red","purple","grey","orange")),
genome=list("C.CK" = 189.51,"C.NPK"=188,"GC.CK"=186.11,
"GC.NPK"=191.51,"Alphaproteobacteria"=70.16,"Betaproteobacteria"=23.51,"Gammaproteobacteria"=25.51,
"Deltaproteobacteria"=23.28,"Acidobacteria"=53.62,"Actinobacteria"=72.33,"Bacteroidetes"=22.41,
"Chloroflexi"=15.08,"Firmicutes"=10.72,"Gemmatimonadetes"=26.37,"Planctomycetes"=19.26,"Thaumarchaeota"=6.15,
"Verrucomicrobia"=8.3,"Ascomycota"=159.41,"Basidiomycota"=79.73,"Zygomycota"=139.29 ),outerRadius = 217,
genomeLabelDisplay = FALSE,CHORDMouseEvent =TRUE,CHORDMouseOverDisplay = TRUE)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosChord().
CircosChord.p() can display the relationship between gene, exon, protein, etc. (Using start/end position as chord path)
library(interacCircos)
chordData=chord.pExample
Circos(moduleList=CircosChord.p('CHORD01', data = chordData,radius= 216,opacity=0.67,
color="black"), innerRadius = 217, outerRadius = 237,genomeLabelDisplay = TRUE,zoom=FALSE)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosChord.p().
CircosChord.p() now is not compatible with zoom and compare function of BioCircos.js/NG-Circos
CircosCnv() can display the CNV data, etc.
library(interacCircos)
cnvData=cnvExample
Circos(moduleList=CircosCnv('Cnv01',maxRadius =175, minRadius =116,
data =cnvData,width=2,color = "#4876FF",animationDisplay = TRUE,
animationType = "linear",animationTime = 500)+
CircosBackground("bg01",minRadius = 116,maxRadius = 175,
fillColors = "#F2F2F2",axisShow = TRUE),CNVMouseOverDisplay = TRUE,
CNVMouseOverTooltipsSetting = "style1",CNVMouseOutDisplay = TRUE,
CNVMouseOutColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosCnv().
CircosGene() can display the gene profile, transcript profile, RNA profile, etc.
library(interacCircos)
geneData=geneExample
Circos(moduleList=CircosGene('Gene01', outerRadius = 195, innerRadius = 180,
data=geneData,arrowGap = 10, arrowColor = "black",arrowSize = "12px",
cdsColor = "#1e77b3",cdsStrokeColor = "#1e77b3",cdsStrokeWidth= 0,
utrWidth= 0,utrColor= "#fe7f0e",utrStrokeColor= "#fe7f0e",
animationDisplay = TRUE),genome =list("EGFR"=1000),outerRadius = 220,
GENEMouseOverDisplay = TRUE,GENEMouseOverTooltipsSetting = "style1",
GENEMouseOutDisplay = TRUE,GENEMouseOutColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosGene().
CircosHeatmap() can display the gene expression, probe signal, etc.
library(interacCircos)
heatmapData=heatmapExample
Circos(moduleList=CircosHeatmap('Heatmap01', maxRadius= 180, minRadius = 100,
data=heatmapData,totalLayer = 3,animationDisplay = TRUE),
genome = list("2L"=23011544,"2R"=21146708,"3L"=24543557,"3R"=27905053,
"4"=1351857,"X"=22422827),HEATMAPMouseEvent = TRUE,HEATMAPMouseOverDisplay = TRUE,
HEATMAPMouseOverTooltipsSetting = "style1",HEATMAPMouseOutDisplay = TRUE,
HEATMAPMouseOutColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosHeatmap().
CircosHistogram() can display the gene expression, probe signal, etc.
library(interacCircos)
histogramData=histogramExample
Circos(moduleList=CircosHistogram('HISTOGRAM01', data = histogramData,
fillColor= "#ff7f0e",maxRadius = 210,minRadius = 175,
animationDisplay = TRUE),genome=list("2L"=23011544,"2R"=21146708,"3L"=24543557,
"3R"= 27905053,"X"=22422827,"4"=1351857),outerRadius = 220,
HISTOGRAMMouseOverDisplay = TRUE,HISTOGRAMMouseOverTooltipsSetting = "style1",
HISTOGRAMMouseOutDisplay = TRUE,HISTOGRAMMouseOutColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosHistogram().
CircosLegend() can display legend easily for better explain each element.
library(interacCircos)
legend1 = list(type= "circle", color="#1E77B4",opacity="1.0",
circleSize="8",text= "C.CK", textSize= "14",textWeight="normal")
legend2 = list(type= "circle", color="#AEC7E8",opacity="1.0",
circleSize="8",text= "C.NPK", textSize= "14",textWeight="normal")
Circos(moduleList=CircosLegend('legend01', title = "legend",
data=list(legend1,legend2),size = 20))
Note : No data should be input. The details for parameters are available by ?CircosLegend().
CircosLine() can display the gene expression, r2 value, etc.
library(interacCircos)
lineData=lineExample
Circos(moduleList=CircosLine('LINE01', data = lineData,
maxRadius=200,minRadius=150,color= "#ff0031",
animationDisplay = TRUE,animationTime = 4000,animationDelay = 100)+
CircosBackground('BG01',minRadius = 205,maxRadius = 150),
LINEMouseOverDisplay = TRUE,LINEMouseOverTooltipsSetting = "style1",
LINEMouseOutDisplay = TRUE,LINEMouseOutLineStrokeColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosLine().
CircosLink() can display the gene fusions, interaction, SV data, etc.
library(interacCircos)
linkData=linkExample
Circos(moduleList=CircosLink('LINK01', data = linkData,
LinkRadius= 140,fillColor= "#9e9ac6",width= 2,
axisPad= 3,labelPad=8,animationDisplay=TRUE,
animationDirection="1to2",animationType= "linear" ),
LINKMouseOverTooltipsSetting = "style1",LINKMouseOverDisplay=TRUE,
LINKMouseOutDisplay = TRUE)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosLink().
CircosLollipop() can display the somatic mutation, germline variantion, etc.
library(interacCircos)
lollipopData=lollipopExample
arcData=arcExample
Circos(moduleList=CircosLollipop('Lollipop01',
data=lollipopData, fillColor="#9400D3",circleSize= 6,
strokeColor= "#999999", strokeWidth= "1px",
animationDisplay=TRUE, lineWidth= 2,realStart= 101219350)+
CircosArc('Arc01', outerRadius = 212, innerRadius = 224, data=arcData),
genome=list("EGFR"=1211),outerRadius = 220,genomeFillColor = c("grey"),
LOLLIPOPMouseOverDisplay = TRUE,LOLLIPOPMouseOverTooltipsSetting = "style1",
LOLLIPOPMouseOutDisplay = TRUE,LOLLIPOPMouseOutColor = NULL,
LOLLIPOPMouseOutCircleSize = 6,LOLLIPOPMouseOutCircleStrokeWidth = "1px",
LOLLIPOPMouseOutCircleStrokeColor = "#999999")
Note : The data is input through the data parameter. The details for parameters are available by ?CircosLollipop().
CircosScatter() can display the SNP without value, Genes data, etc.
library(interacCircos)
scatterData=scatterExample
Circos(moduleList=CircosScatter('SCATTER01', data = scatterData,
radius=180,innerCircleColor= "#3d6390",outerCircleColor= "#99cafe",
random_data= 40,animationDisplay = TRUE,animationDelay = 10,
animationTime = 1000,animationType = "linear"),SCATTERMouseOverDisplay = TRUE,
SCATTERMouseOverTooltipsSetting = "style1",SCATTERMouseOutDisplay = TRUE,
SCATTERMouseOutColor = NULL,SCATTERMouseOutCircleStrokeColor ="#99cafe",
SCATTERMouseOutCircleStrokeWidth = 6)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosScatter().
CircosSnp() can display the SNP density data, GWAS data, etc.
library(interacCircos)
snpData=snpExample
Circos(moduleList=CircosSnp('SNP01', minRadius =150,
maxRadius = 190, data = snpExample,SNPFillColor= "#9ACD32",
circleSize= 2, SNPAxisColor= "#B8B8B8", SNPAxisWidth= 0.5,
animationDisplay=TRUE,animationTime= 2000, animationDelay= 0,
animationType= "linear") +
CircosBackground('BG01',minRadius = 145,maxRadius = 200),
SNPMouseOverDisplay = TRUE,SNPMouseOverTooltipsSetting = "style1",
SNPMouseOutDisplay = TRUE,SNPMouseOutColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosSnp().
CircosText() can display annotation text at specific elements.
library(interacCircos)
Circos(moduleList=CircosText('text01', 'Annotation',
color = '#DD2222', x = -40,animationDisplay = TRUE,
animationInitialPositionX = 200))
Note : No data should be input. The details for parameters are available by ?CircosText().
CircosWig() can display the gene expression data, read depth etc.
library(interacCircos)
wigData=wigExample
Circos(moduleList=CircosWig('WIG01', data = wigData,
maxRadius= 200,minRadius= 150,strokeColor= "darkblue",
color= "lightblue",strokeType= "cardinal",
animationDisplay = TRUE,animationTime = 3000,animationType="linear")+
CircosBackground('BG01',minRadius = 205,maxRadius = 150),
genome=list("chr8"=1000),outerRadius = 220,WIGMouseOverDisplay = TRUE,
WIGMouseOverTooltipsSetting = "style1",WIGMouseOutDisplay = TRUE,
WIGMouseOutFillColor = NULL)
Note : The data is input through the data parameter. The details for parameters are available by ?CircosWig().