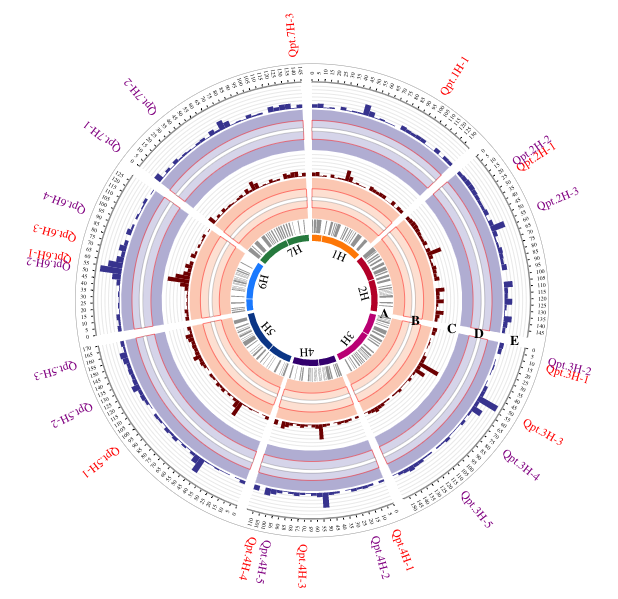
An interactive Circos plot indicating QTL involved in net blotch resistance and displayed using histogram, arc, link, background, text function.
data.frame in R is a table or a two-dimensional array-like structure in which each column contains values of one variable and each row contains one set of values from each column.
The details for data.frame is available here
It is because that most of R packages are supporting this type of data structure, such as tidyverse, reshape2...
And meanwhile, data.frame can be easily transformed to other structure, such as matrix, array, list...
| chr | start | end | color | des | link | html |
|---|---|---|---|---|---|---|
| EGFR | 57 | 167 | #FFBB77 | Recep_L_domain | NA | |
| EGFR | 185 | 338 | #FF7F0E | Furin-like_domain | https://www.intogen.org/search?gene=EGFR | NA |
| EGFR | 361 | 480 | #FFBB77 | Recep_L_domain | https://www.intogen.org/search?gene=EGFR | NA |
| EGFR | 505 | 636 | #1F77B4 | GF_recep_IV_domain | NA | |
| EGFR | 713 | 965 | #AEC6E7 | Pkinase_Tyr_domain | NA |
Full example of data can be found through ?arcExample
No data should be input to CircosAuxLine()
No data should be input to CircosBackground()
| chr | start | end | name | value | layer | color | html |
|---|---|---|---|---|---|---|---|
| 2L | 1 | 1011544 | TP53 | 0.8 | 1 | NA | NA |
| 2L | 1011548 | 2011544 | TP53 | 0.3 | 1 | NA | NA |
| ... | ... | ... | ... | ... | ... | ... | ... |
| X | 20011548 | 21146708 | TP53 | 0.49 | 1 | NA | NA |
| X | 21011548 | 22422827 | TP53 | 0.83 | 1 | NA | NA |
Full example of data can be found through ?bubbleExample
| C.CK | C.NPK | ... | Basidiomycota | Zygomycota |
|---|---|---|---|---|
| 0 | 0 | ... | 24.75 | 18.49 |
| 0 | 0 | ... | 24.18 | 29.3 |
| ... | ... | ... | ... | ... |
| 24.75 | 24.18 | ... | 0 | 0 |
| 18.49 | 29.3 | ... | 0 | 0 |
Full example of data can be found through ?chordExample
| source_chr | source_start | source_end | target_chr | target_start | target_end |
|---|---|---|---|---|---|
| 19 | 22186054 | 26186054 | 17 | 31478117 | 35478117 |
| 4 | 74807187 | 78807187 | 1 | 89852878 | 93852878 |
| ... | ... | ... | ... | ... | ... |
| 17 | 31478117 | 35478117 | 1 | 89852849 | 93852849 |
| 17 | 31478114 | 31478114 | X | 106297713 | 110297713 |
Full example of data can be found through ?chord.pExample
| chr | start | end | value | link | color | html |
|---|---|---|---|---|---|---|
| 1 | 764788 | 87109267 | 2.5 | NA | NA | NA |
| 1 | 87109268 | 120217058 | 2.0 | NA | NA | NA |
| ... | ... | ... | ... | ... | ... | ... |
| 3 | 125251487 | 125456654 | 5 | NA | NA | NA |
| 3 | 125456655 | 127048011 | 4 | NA | NA | NA |
Full example of data can be found through ?cnvExample
| chr | strand | start | end | type | name | link | html |
|---|---|---|---|---|---|---|---|
| EGFR | - | 1 | 250 | gene | EGFR | https://www.intogen.org/search?gene=EGFR | NA |
| EGFR | - | 1 | 30 | utr | EGFR | https://www.intogen.org/search?gene=EGFR | NA |
| ... | ... | ... | ... | ... | ... | ... | ... |
| EGFR | - | 920 | 935 | cds | EGFR | https://www.intogen.org/search?gene=EGFR | NA |
| EGFR | - | 935 | 950 | utr | EGFR | https://www.intogen.org/search?gene=EGFR | NA |
Full example of data can be found through ?geneExample
| chr | start | end | name | value | layer | html | |
|---|---|---|---|---|---|---|---|
| 2L | 1 | 1011544 | TP53 | 0.8 | 1 | NA | NA |
| 2L | 1011548 | 2011544 | TP53 | 0.3 | 1 | NA | NA |
| ... | ... | ... | ... | ... | ... | ... | ... |
| X | 20011548 | 21146708 | TP53 | 0.49 | 1 | NA | NA |
| X | 21011548 | 22422827 | TP53 | 0.83 | 1 | NA | NA |
Full example of data can be found through ?heatmapExample
| chr | start | end | name | value | layer | html | |
|---|---|---|---|---|---|---|---|
| 2L | 1 | 1011544 | TP53 | 0.8 | 1 | NA | NA |
| 2L | 1011548 | 2011544 | TP53 | 0.3 | 1 | NA | NA |
| ... | ... | ... | ... | ... | ... | ... | ... |
| X | 20011548 | 21146708 | TP53 | 0.49 | 1 | NA | NA |
| X | 21011548 | 22422827 | TP53 | 0.83 | 1 | NA | NA |
Full example of data can be found through ?histogramExample
No data should be input to CircosLegend()
| chr | pos | value | des | html |
|---|---|---|---|---|
| 1 | 813468 | 0.15 | NA | NA |
| 1 | 3383745 | 0.122385 | NA | NA |
| ... | ... | ... | ... | ... |
| 7 | 141390027 | 0.0922775 | NA | NA |
| 7 | 146503088 | 0.1907651 | NA | NA |
Full example of data can be found through ?lineExample
| fusion | g1chr | g1start | g1end | g1name | g2chr | g2start | g2end | g2name | link | html |
|---|---|---|---|---|---|---|---|---|---|---|
| FGFR3--TACC3 | 4 | 1795662 | 1808986 | FGFR3 | 4 | 1723217 | 1746905 | TACC3 | NA | NA |
| CCDC6--RET | 10 | 43595894 | 43623714 | RET | 10 | 61552678 | 61666182 | TACC3 | NA | NA |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ABL1--BCR | 9 | 133589707 | 133761067 | ABL1 | 22 | 23523148 | 23657706 | BCR | NA | NA |
| SND1--BRAF | 7 | 127292043 | 127732659 | SND1 | 7 | 140434400 | 140624503 | BRAF | NA | NA |
Full example of data can be found through ?linkExample
| protein | chr | pos | strand | CancerTypeNumber | AA_pos | AA_change | Consequence | color | type | link | html |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EGFR | 7 | 101219410 | + | 2 | AA_858 | L/R | missense_variant | #FEFF00 | Hetero | NA | |
| EGFR | 7 | 101219417 | + | 2 | AA_858 | L/R | missense_variant | #FEFF00 | Homo | NA | |
| EGFR | 7 | 101219425 | + | 1 | AA_858 | L/R | missense_variant | #FEFF00 | Homo | NA | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
Full example of data can be found through ?lollipopExample
| chr | start | end | name | des | link | html |
|---|---|---|---|---|---|---|
| 12 | 125550131 | 125626772 | AACS | liver | NA | NA |
| 9 | 107546599 | 107665960 | ABCA1 | colorectal | NA | NA |
| 16 | 2326678 | 2376467 | ABCA3 | breast | NA | NA |
| ... | ... | ... | ... | ... | ... | ... |
Full example of data can be found through ?scatterExample
| chr | pos | value | des | color | r2value | link | index | image | html |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 813468 | 0.714061 | GWAS_SNP | NA | NA | NA | NA | NA | NA |
| 1 | 3383745 | 0.22385 | GWAS_SNP | NA | NA | NA | NA | NA | NA |
| 1 | 6157646 | 0.238643 | GWAS_SNP | NA | NA | NA | NA | NA | NA |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
Full example of data can be found through ?snpExample
No data should be input to CircosText()
| chr | pos | value | des | html |
|---|---|---|---|---|
| chr8 | 0.024557353699565337 | 5 | NA | NA |
| chr8 | 0.5157044276908721 | 5 | NA | NA |
| chr8 | 1.0068515016821786 | 4 | NA | NA |
| ... | ... | ... | ... | ... |
Full example of data can be found through ?wigExample